
Jong-In Park, PhD
Professor; Eminent Scholar
Locations
- Biochemistry
BSB 357
Contact Information
General Interests
Education
PhD, University of New South Wales, Sydney, Australia, 2000
MA, Yonsei University, Seoul, Korea, 1990
BA, Yonsei University, Seoul, Korea, 1988
Biography
Dr. Park earned his Bachelor’s and Master’s degrees in Biochemistry and worked for the pharmaceutical branch of SAMSUNG, Inc until he decided to pursue an academic career. Dr. Park received his PhD degree in Biochemistry and Molecular Genetics for studies of Ras-regulated stress responses in yeast. He then conducted postdoctoral research on the role of the MAP kinase pathways for oncogenic Ras, Raf, and receptor tyrosine kinases in human cancer. Upon completing his training, Dr. Park joined the faculty of the Biochemistry Department at the Medical College of Wisconsin in 2006. Since then, Dr. Park has been studying oncogenic signaling and metabolic pathways in different tumors with the support from various funding agencies, including the NIH-National Cancer Institute, the American Cancer Society, and the Department of Defense.
In addition to basic science research, Dr. Park also conducts clinical cancer research. For example, since 2014, he has been participating in the NCI-MATCH Precision Medicine Cancer Trial as the Translational Chair of the Dabrafenib & Trametinib combination therapy arm, which targets BRAF-driven cancer. Dr. Park is a former recipient of the ACS Research Scholar award, the DOD New Investigator award, and the FAMRI Young Investigator award. At MCW, Dr. Park received multiple recognitions as Outstanding Medical Student Teacher and Outstanding Graduate Student Teacher.
Research Experience
- Cell Cycle
- Cell Death
- Drug Resistance, Neoplasm
- Gene Expression Regulation, Neoplastic
- MAP Kinase Signaling System
- Melanoma
- Metabolism
- Mitochondria
- Molecular Chaperones
- Pancreatic Neoplasms
- Precision Medicine
- Thyroid Neoplasms
Research Interests
Oncogenic transformation requires reprogramming in signaling and metabolism, which inevitably causes cellular stress that places pressure on transforming cells to develop stress tolerance mechanisms. As such, malignant tumors might have successfully developed a protective mechanism and, if identified, this mechanism may be targeted for therapy. An important goal of Park lab research is to elucidate the molecular mechanisms underlying these events and to translate the knowledge into an advanced therapeutic strategy.
Dr. Park’s research projects include:
Studying the role of mortalin in MEK/ERK-activated tumor cells
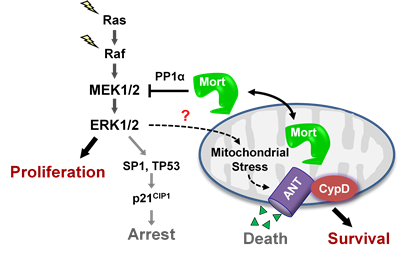 The MEK/ERK pathway is a key effector of the oncogenic BRAF, KRAS, and receptor tyrosine kinases, and its deregulation is a central signature of many epithelial cancers. We recently demonstrated that MEK/ERK deregulation puts cells at risk of mitochondrial cell death, but that mortalin, a mitochondrial chaperone, can counteract this risk. Briefly, mortalin can determine the live/die decision in MEK/ERK-dependent tumor cells by regulating a mitochondrial death machinery that consists of ANT, a mitochondrial channel that controls bioenergetic homeostasis, and CypD, a chaperone that functions as the gatekeeper of the mitochondrial permeability transition pore (view article PMID: 32156782; view article PMID: 32291414). Current studies in Park lab focus on elucidating the nature of mitochondrial stress associated with overactive MEK/ERK signaling in tumor cells and on identifying the molecular mechanisms by which mortalin protects tumor cells from this stress.
The MEK/ERK pathway is a key effector of the oncogenic BRAF, KRAS, and receptor tyrosine kinases, and its deregulation is a central signature of many epithelial cancers. We recently demonstrated that MEK/ERK deregulation puts cells at risk of mitochondrial cell death, but that mortalin, a mitochondrial chaperone, can counteract this risk. Briefly, mortalin can determine the live/die decision in MEK/ERK-dependent tumor cells by regulating a mitochondrial death machinery that consists of ANT, a mitochondrial channel that controls bioenergetic homeostasis, and CypD, a chaperone that functions as the gatekeeper of the mitochondrial permeability transition pore (view article PMID: 32156782; view article PMID: 32291414). Current studies in Park lab focus on elucidating the nature of mitochondrial stress associated with overactive MEK/ERK signaling in tumor cells and on identifying the molecular mechanisms by which mortalin protects tumor cells from this stress.
Targeting mitochondrial metabolism in RET-mutant cancers
Somatic as well as inherited mutations in the RET receptor tyrosine kinase are a key etiological factor in cancers, including thyroid cancer. For example, inherited RET mutations are an important prognostic marker for the multiple endocrine neoplasia type 2 (MEN2) syndrome, in which medullary thyroid cancer is a key pathological presentation. As a member of the American Cancer Society MEN2 consortium, we recently demonstrated that mitochondrial bioenergetics is affected by RET inhibition in medullary thyroid tumor cells and that the mitochondrial activity altered upon RET inhibition can be exploited for the design of a combination therapy with mitochondria targeted agents (view article PMID: 28475408). Our current research focuses on further developing this concept in different tumors.
Participating in the NCI-MATCH Precision Medicine Cancer Trial
This clinical trial is a “genotype to phenotype” phase II study. An important goal of this study is to identify the features of various tumor types with the same mutation that cause them to either respond to or resist treatment with a targeted therapy (view article PMID: 32758030). Find more information at the National Cancer Institute.
Publications
-
ERK1/2 interaction with DHPS regulates eIF5A deoxyhypusination independently of ERK kinase activity.
(Becker AE, Kochanowski P, Wu PK, Wątor E, Chen W, Guchhait K, Biela AP, Grudnik P, Park JI.) Cell Rep. 2024 Oct 22;43(10):114831 PMID: 39392755 PMCID: PMC11544350 SCOPUS ID: 2-s2.0-85207351264 10/11/2024
-
(Chen W, Song YS, Lee HS, Lin CW, Lee J, Kang YE, Kim SK, Kim SY, Park YJ, Park JI.) Oncogene. 2024 Jul;43(31):2431-2446 PMID: 38937602 PMCID: PMC11629884 SCOPUS ID: 2-s2.0-85197858420 06/28/2024
-
(Leung PY, Chen W, Sari AN, Sitaram P, Wu PK, Tsai S, Park JI.) World J Gastroenterol. 2024 Feb 21;30(7):714-727 PMID: 38515951 PMCID: PMC10950623 SCOPUS ID: 2-s2.0-85185579019 03/22/2024
-
(Chen W, Dream S, Leung PY, Wu PK, Wong S, Park JI.) NPJ Precis Oncol. 2024 Feb 20;8(1):39 PMID: 38378752 PMCID: PMC10879150 SCOPUS ID: 2-s2.0-85185513016 02/21/2024
-
Tumor Cell Resistance to the Inhibition of BRAF and MEK1/2.
(Chen W, Park JI.) Int J Mol Sci. 2023 Oct 02;24(19) PMID: 37834284 PMCID: PMC10573597 SCOPUS ID: 2-s2.0-85174308251 10/14/2023
-
(Kim J, Zimmerman MA, Shin WY, Boettcher BT, Lee JS, Park JI, Ali M, Yang M, Mishra J, Hagen CE, McGraw JE, Mathison A, Woehlck HJ, Lomberk G, Camara AKS, Urrutia RA, Stowe DF, Hong JC.) Ann Surg. 2023 Feb 01;277(2):e366-e375 PMID: 34387201 PMCID: PMC8840998 SCOPUS ID: 2-s2.0-85146161896 08/14/2021
-
(Kim J, Hong SK, Yang Y, Lee A, Hoffmeister KM, Gantner BN, Park JI.) Front Transplant. 2023;2:1215182 PMID: 38993858 PMCID: PMC11235240 SCOPUS ID: 2-s2.0-85204385238 07/12/2024
-
(Park JI.) International Journal of Molecular Sciences. June 2023;24(11) SCOPUS ID: 2-s2.0-85161598318 06/01/2023
-
Editorial: The role of mortalin in biology and disease
(Park JI.) Frontiers in Cell and Developmental Biology. 2023;11 SCOPUS ID: 2-s2.0-85153495202 01/01/2023
-
Analogs of the Heat Shock Protein 70 Inhibitor MKT-077 Suppress Medullary Thyroid Carcinoma Cells.
(Hong SK, Starenki D, Johnson OT, Gestwicki JE, Park JI.) Int J Mol Sci. 2022 Jan 19;23(3) PMID: 35162987 PMCID: PMC8835675 SCOPUS ID: 2-s2.0-85122962789 02/16/2022
-
eIF5A-Independent Role of DHPS in p21CIP1 and Cell Fate Regulation.
(Becker AE, Wu PK, Park JI.) Int J Mol Sci. 2021 Dec 07;22(24) PMID: 34947982 PMCID: PMC8707118 SCOPUS ID: 2-s2.0-85120635476 12/25/2021
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.
(Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, Adamopoulos IE, Adeli K, Adolph TE, Adornetto A, Aflaki E, Agam G, Agarwal A, Aggarwal BB, Agnello M, Agostinis P, Agrewala JN, Agrotis A, Aguilar PV, Ahmad ST, Ahmed ZM, Ahumada-Castro U, Aits S, Aizawa S, Akkoc Y, Akoumianaki T, Akpinar HA, Al-Abd AM, Al-Akra L, Al-Gharaibeh A, Alaoui-Jamali MA, Alberti S, Alcocer-Gómez E, Alessandri C, Ali M, Alim Al-Bari MA, Aliwaini S, Alizadeh J, Almacellas E, Almasan A, Alonso A, Alonso GD, Altan-Bonnet N, Altieri DC, Álvarez ÉMC, Alves S, Alves da Costa C, Alzaharna MM, Amadio M, Amantini C, Amaral C, Ambrosio S, Amer AO, Ammanathan V, An Z, Andersen SU, Andrabi SA, Andrade-Silva M, Andres AM, Angelini S, Ann D, Anozie UC, Ansari MY, Antas P, Antebi A, Antón Z, Anwar T, Apetoh L, Apostolova N, Araki T, Araki Y, Arasaki K, Araújo WL, Araya J, Arden C, Arévalo MA, Arguelles S, Arias E, Arikkath J, Arimoto H, Ariosa AR, Armstrong-James D, Arnauné-Pelloquin L, Aroca A, Arroyo DS, Arsov I, Artero R, Asaro DML, Aschner M, Ashrafizadeh M, Ashur-Fabian O, Atanasov AG, Au AK, Auberger P, Auner HW, Aurelian L, Autelli R, Avagliano L, Ávalos Y, Aveic S, Aveleira CA, Avin-Wittenberg T, Aydin Y, Ayton S, Ayyadevara S, Azzopardi M, Baba M, Backer JM, Backues SK, Bae DH, Bae ON, Bae SH, Baehrecke EH, Baek A, Baek SH, Baek SH, Bagetta G, Bagniewska-Zadworna A, Bai H, Bai J, Bai X, Bai Y, Bairagi N, Baksi S, Balbi T, Baldari CT, Balduini W, Ballabio A, Ballester M, Balazadeh S, Balzan R, Bandopadhyay R, Banerjee S, Banerjee S, Bánréti Á, Bao Y, Baptista MS, Baracca A, Barbati C, Bargiela A, Barilà D, Barlow PG, Barmada SJ, Barreiro E, Barreto GE, Bartek J, Bartel B, Bartolome A, Barve GR, Basagoudanavar SH, Bassham DC, Bast RC Jr, Basu A, Batoko H, Batten I, Baulieu EE, Baumgarner BL, Bayry J, Beale R, Beau I, Beaumatin F, Bechara LRG, Beck GR Jr, Beers MF, Begun J, Behrends C, Behrens GMN, Bei R, Bejarano E, Bel S, Behl C, Belaid A, Belgareh-Touzé N, Bellarosa C, Belleudi F, Belló Pérez M, Bello-Morales R, Beltran JSO, Beltran S, Benbrook DM, Bendorius M, Benitez BA, Benito-Cuesta I, Bensalem J, Berchtold MW, Berezowska S, Bergamaschi D, Bergami M, Bergmann A, Berliocchi L, Berlioz-Torrent C, Bernard A, Berthoux L, Besirli CG, Besteiro S, Betin VM, Beyaert R, Bezbradica JS, Bhaskar K, Bhatia-Kissova I, Bhattacharya R, Bhattacharya S, Bhattacharyya S, Bhuiyan MS, Bhutia SK, Bi L, Bi X, Biden TJ, Bijian K, Billes VA, Binart N, Bincoletto C, Birgisdottir AB, Bjorkoy G, Blanco G, Blas-Garcia A, Blasiak J, Blomgran R, Blomgren K, Blum JS, Boada-Romero E, Boban M, Boesze-Battaglia K, Boeuf P, Boland B, Bomont P, Bonaldo P, Bonam SR, Bonfili L, Bonifacino JS, Boone BA, Bootman MD, Bordi M, Borner C, Bornhauser BC, Borthakur G, Bosch J, Bose S, Botana LM, Botas J, Boulanger CM, Boulton ME, Bourdenx M, Bourgeois B, Bourke NM, Bousquet G, Boya P, Bozhkov PV, Bozi LHM, Bozkurt TO, Brackney DE, Brandts CH, Braun RJ, Braus GH, Bravo-Sagua R, Bravo-San Pedro JM, Brest P, Bringer MA, Briones-Herrera A, Broaddus VC, Brodersen P, Brodsky JL, Brody SL, Bronson PG, Bronstein JM, Brown CN, Brown RE, Brum PC, Brumell JH, Brunetti-Pierri N, Bruno D, Bryson-Richardson RJ, Bucci C, Buchrieser C, Bueno M, Buitrago-Molina LE, Buraschi S, Buch S, Buchan JR, Buckingham EM, Budak H, Budini M, Bultynck G, Burada F, Burgoyne JR, Burón MI, Bustos V, Büttner S, Butturini E, Byrd A, Cabas I, Cabrera-Benitez S, Cadwell K, Cai J, Cai L, Cai Q, Cairó M, Calbet JA, Caldwell GA, Caldwell KA, Call JA, Calvani R, Calvo AC, Calvo-Rubio Barrera M, Camara NO, Camonis JH, Camougrand N, Campanella M, Campbell EM, Campbell-Valois FX, Campello S, Campesi I, Campos JC, Camuzard O, Cancino J, Candido de Almeida D, Canesi L, Caniggia I, Canonico B, Cantí C, Cao B, Caraglia M, Caramés B, Carchman EH, Cardenal-Muñoz E, Cardenas C, Cardenas L, Cardoso SM, Carew JS, Carle GF, Carleton G, Carloni S, Carmo.) Autophagy. 2021 Jan;17(1):1-382 PMID: 33634751 PMCID: PMC7996087 SCOPUS ID: 2-s2.0-85102619204 02/27/2021

