
Brian A. Link, PhD
Professor
Locations
- Cell Biology, Neurobiology and Anatomy
Contact Information
Education
PhD, Oregon Health & Sciences University, 1997
BS, University of California, San Diego, CA, 1991
Research Interests
The overarching research goal of the Link lab is to study the cellular basis of signaling and the role in development and relationships to disease processes. We primarily use zebrafish for our studies, combining imaging based technologies and genome editing. As part of this research we have developed tools to monitor and manipulate basic cellular processes such as endocytosis, vesicle trafficking and nuclear dynamics. In addition, we have created transgenic animals to investigate and quantitate signaling networks including Notch, BMP/Smad, and Hippo-Yap/Taz. There are several areas of research we are currently focused. One goal is to understand the cell biology and signaling events critical for ocular development and disease. Currently we are studying how endocytosis and polarized vesicle trafficking affect signaling, primarily Notch, Wnt and Hippo-Yap/Taz pathways, in neuroepithelia and how these pathways interact to regulate neurogenesis. Ancillary to these studies, we are characterizing the role of Yap/Taz activity in retinal pigment epithelial development and maintenance. For ocular disease studies, we are characterizing the cellular mechanisms underlying pathology in several eye disorders including PhR degenerations, glaucomas, and myopia. Integrated with all these research goals, the Link lab emphasizes collaborative research and career training for students and post-docs.
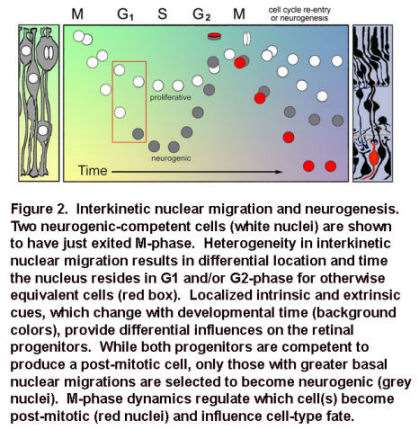
Movie 1. In vivo confocal time-lapse movie shows heterogeneity of interkinetic nuclear migration in retinal progenitors from 24 hpf.
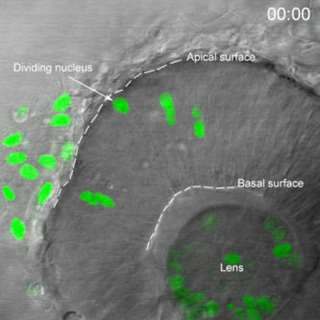
Images are composed of compressed z-stacks of H2A-GFP fluorescence overlaid with single mid-plane bright-field images. Apical and basal surfaces are outlined with dashed white lines in the first and last frame and an arrow indicates a cell undergoing mitosis at the apical surface. The movie plays at 6 frames per second covering 12 hr of development time (Baye and Link, 2007).
Specifically, neuroepithelia with deep basal nuclear migrations and shorter cell cycles are biased to produce neurons, rather than give rise to daughter cells that re-enter the cell cycle. The lab is currently exploring the signals and mechanisms underlying these relationships.
Complex gene interactions underlying glaucoma-phenotypes
The overall goal of these studies is to understand the relationships between genes that contribute to glaucoma-phenotypes. The glaucomas are a heterogeneous group of ocular disorders characterized by retinal ganglion cell death, optic nerve damage and visual field loss. Elevated intraocular pressure is a principal risk factor for this neurodegenerative disease. The majority and perhaps all forms of glaucoma are complex – requiring the interaction of multiple genes. While several genes that contribute to glaucoma have been identified, many additional loci remain unknown. We are using zebrafish to identify and study genes which promote glaucoma-phenotypes (Figure 3).
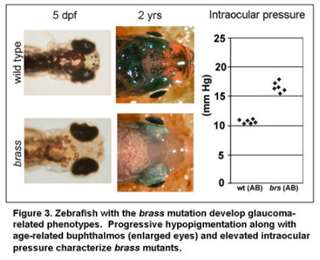
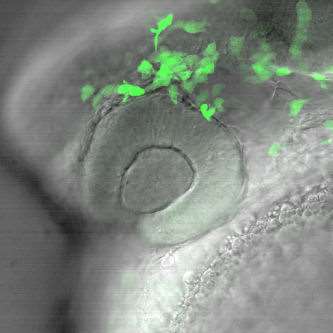
In particular, we are focusing on the complex-gene interactions that promote retinal ganglion cell death in the context of elevated intraocular pressure. Additional projects are focused on genes that regulate the development of glaucoma-relevant ocular tissues, as such genes have previously been shown to impact glaucoma (Movie 2).
Movie 2. In vivo confocal time-lapse movie showing migration of neural crest cells into the periocular region of the eye. These cells, along with head mesoderm, contribute to the structures of the anterior segment that regulate intraocular pressure. Cells were labeled with the foxd3:GFP transgene (Gilmour et al. Neuron 34:577-88, 2002)
Publications
-
(Nonarath HJT, Jackson MA, Penoske RM, Zahrt TC, Price NPJ, Link BA.) J Antibiot (Tokyo). 2024 Apr;77(4):245-256 PMID: 38238588 PMCID: PMC11403873 SCOPUS ID: 2-s2.0-85182630226 01/19/2024
-
(Eisa-Beygi S, Hu MM, Kumar SN, Jeffery BE, Collery RF, Vo NJ, Lamichanne BS, Yost HJ, Veldman MB, Link BA.) Arterioscler Thromb Vasc Biol. 2023 Jul;43(7):e231-e237 PMID: 37128914 PMCID: PMC10330147 SCOPUS ID: 2-s2.0-85163499929 05/02/2023
-
Myofibroblast Ccn3 is regulated by Yap and Wwtr1 and contributes to adverse cardiac outcomes.
(Flinn MA, Alvarez-Argote S, Knas MC, Almeida VA, Paddock SJ, Zhou X, Buddell T, Jamal A, Taylor R, Liu P, Drnevich J, Patterson M, Link BA, O'Meara CC.) Front Cardiovasc Med. 2023;10:1142612 PMID: 36998974 PMCID: PMC10043314 04/01/2023
-
Endothelial cilia dysfunction in pathogenesis of hereditary hemorrhagic telangiectasia.
(Eisa-Beygi S, Burrows PE, Link BA.) Front Cell Dev Biol. 2022;10:1037453 PMID: 36438574 PMCID: PMC9686338 11/29/2022
-
Ablation of mpeg+ Macrophages Exacerbates mfrp-Related Hyperopia.
(Brandt ZJ, Collery RF, Besharse JC, Link BA.) Invest Ophthalmol Vis Sci. 2021 Dec 01;62(15):13 PMID: 34913948 PMCID: PMC8684298 SCOPUS ID: 2-s2.0-85122380090 12/17/2021
-
Retinal Gene Therapy for Usher Syndrome: Current Developments, Challenges, and Perspectives.
(Dinculescu A, Link BA, Saperstein DA.) Int Ophthalmol Clin. 2021 Oct 01;61(4):109-124 PMID: 34584048 PMCID: PMC8478317 SCOPUS ID: 2-s2.0-85117357508 09/30/2021
-
(Eisa-Beygi S, Vo NJ, Link BA.) Drug Discov Today. 2021 Aug;26(8):1790-1793 PMID: 33358701 SCOPUS ID: 2-s2.0-85098655506 12/29/2020
-
(Clark EM, Link BA.) Mol Biol Cell. 2021 Mar 01;32(5):391-401 PMID: 33439675 PMCID: PMC8098853 SCOPUS ID: 2-s2.0-85101912245 01/14/2021
-
(Eisa-Beygi S, Vo N(, Link BA.) Drug Discovery Today. August 2021;26(8):1790-1793 SCOPUS ID: 2-s2.0-85098655506 08/01/2021
-
Interplay of MPP5a with Rab11 synergistically builds epithelial apical polarity and zonula adherens.
(Hao Y, Zhou Y, Yu Y, Zheng M, Weng K, Kou Z, Liang J, Zhang Q, Tang X, Xu P, Link BA, Yao K, Zou J.) Development. 2020 Nov 19;147(22) PMID: 33060129 SCOPUS ID: 2-s2.0-85096508420 10/17/2020
-
Llgl1 regulates zebrafish cardiac development by mediating Yap stability in cardiomyocytes.
(Flinn MA, Otten C, Brandt ZJ, Bostrom JR, Kenarsary A, Wan TC, Auchampach JA, Abdelilah-Seyfried S, O'Meara CC, Link BA.) Development. 2020 Aug 25;147(16) PMID: 32843528 PMCID: PMC7473637 SCOPUS ID: 2-s2.0-85089923843 08/28/2020
-
(Clark BS, Miesfeld JB, Flinn MA, Collery RF, Link BA.) Front Cell Dev Biol. 2020;8:608112 PMID: 33634099 PMCID: PMC7900515 SCOPUS ID: 2-s2.0-85101223534 02/27/2021
Link Lab


