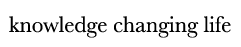Medical College of Wisconsin Liu Laboratory Research
What does it take to regenerate a heart?
Despite wide therapeutic options for treating myocardial infarction (MI), cardiac function recovery in MI patients remains unsatisfactory, urging the need for new strategies to regenerate the heart. Unfortunately, adult mammalian cardiomyocytes (CM) have extremely limited proliferative capacity, and insufficient revascularization post MI fails to restore blood supply of the infarcted area. Alternatives to replenish lost CMs and revascularize the injured area are urgently needed. The long-term goal of the Liu lab is to develop novel strategies for cardiovascular regeneration. To achieve this goal, the lab will explore new approaches of making CMs and blood vessels by elucidating the molecular mechanisms of direct cardiac reprogramming and vascular development and repair.
Direct Cardiac Reprogramming
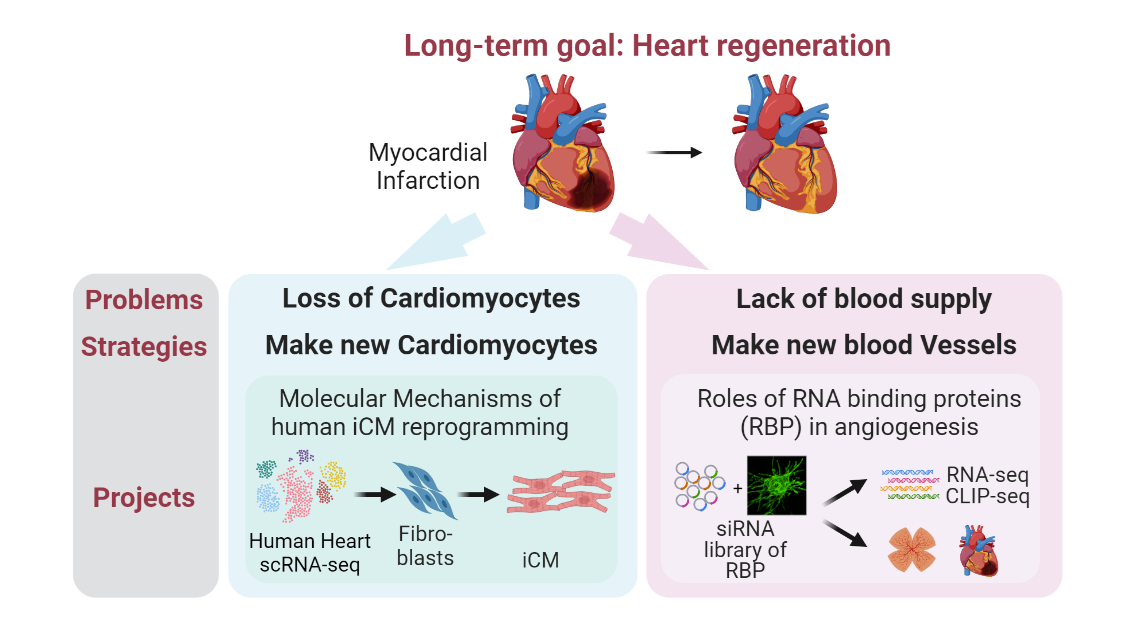
Reprogramming cardiac fibroblasts into CMs with a defined set of transcription factors has emerged as a promising alternative for heart regeneration in the past decade. It attacks two problems in one go - loss of CM and cardiac fibrosis. Previous work in the field mainly focused on mouse reprogramming. Human direct cardiac reprogramming, on the other hand, has substantial clinical relevance, but suffers from longer time and more complex cocktails compared to mouse, suggesting species differences. Therefore, we will combine single-cell RNA-seq analysis with CRISPR screens to identify critical regulators of human cardiac reprogramming. We will then comprehensively characterize whether and how the top candidate from the screen improves the efficiency and quality of reprogramming. This line of research in the lab will shed light on the mechanisms underlying fibroblasts-to-CM fate conversion in human cells and provide with us a better cocktail for potential clinical applications.
Molecular mechanisms of angiogenesis
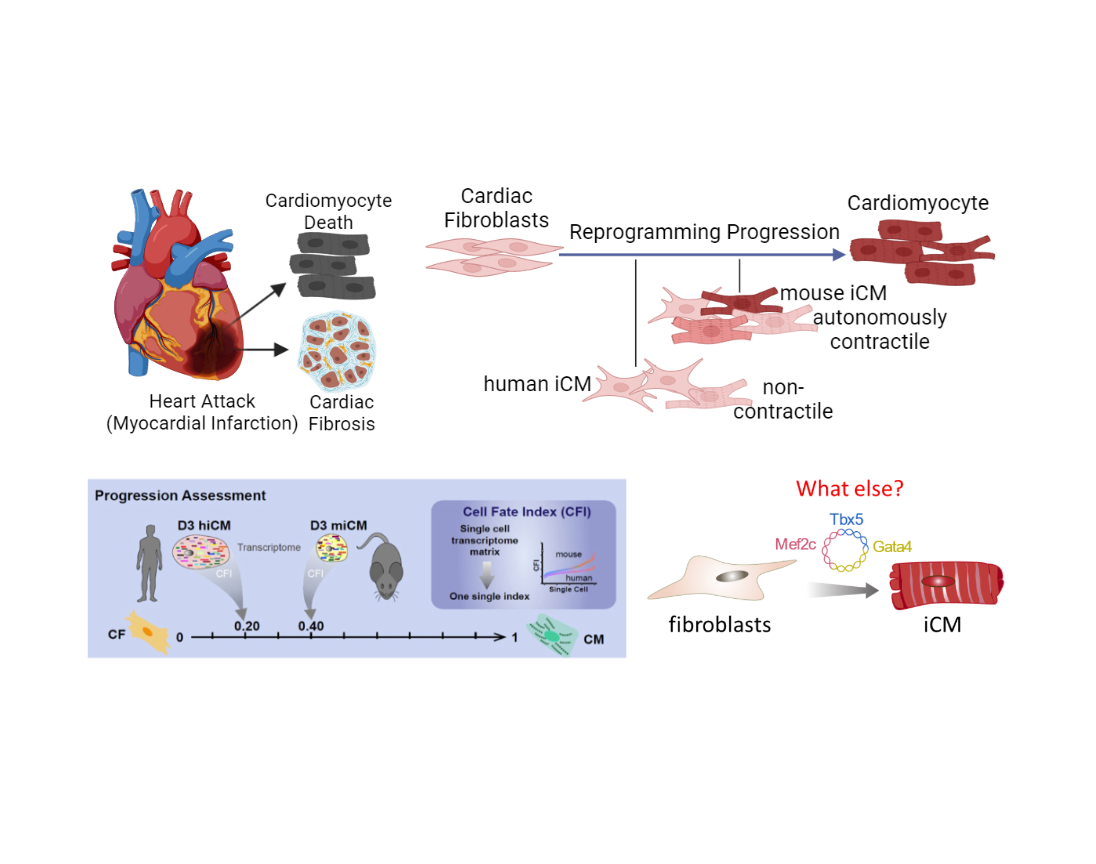
Angiogenesis, the formation of new blood vessels from pre-existing vasculature, is essential for development and regeneration. Research in the past 30 years revealed the critical roles of signaling pathways such as VEGF and their downstream transcriptional events in governing angiogenesis. Nevertheless, angiogenic therapies like VEGFA treatment has been unsuccessful in ischemic heart diseases, underscoring the importance to better understand the molecular basis of angiogenesis. Epigenetic modifications have been implicated in tumor angiogenesis, but whether and how they regulate angiogenesis during development and tissue repair has just started to be tested. Similarly, alternative splicing of key regulators of angiogenesis such as Vegfa and Flt1 was well established but whether and how RNA binding proteins/splicing factors regulate angiogenesis globally is unclear. Therefore, we will combine single-cell omics and loss-of-function screening to identify critical epigenetic factors and RBPs regulating angiogenesis. We will use the neonatal mouse retina model of developmental angiogenesis, coronary artery revascularization post-MI, as well as a 3D in vitro angiogenic sprouting bead assay. We will also use genomic approaches like RNA-seq, CLIP-seq and CUT & RUN to dissect mechanisms. Together, this line of research will illuminate the roles of epigenetic modifiers and RBPs in regulating angiogenesis and fill critical knowledge gaps in the vascular field. It will also identify potential targets for therapeutic intervention of cardiovascular diseases in the future.
Publications
2023
21. Buglak DB, Bougaran P, Kulikauskas MR, Liu Z, Monaghan-Benson E, Gold AL, Marvin AP, Burciu A, Tanke NT, Oatley M, Ricketts SN, Kinghorn K, Johnson BN, Shiau CE, Rogers S, Guilluy C, Bautch VL (2023). Nuclear SUN1 stabilizes endothelial cell junctions via microtubules to regulate blood vessel formation. eLife 12:e83652.2021
20. Liu Z, Ruter DL, Quigley KM, Tanke N, Jiang Y, Bautch VL (2021). Single-Cell RNA Sequencing Reveals Endothelial Cell Transcriptome Heterogeneity under Homeostatic Laminar Flow. Arteriosclerosis, Thrombosis, and Vascular Biology. 2021 Aug 26: ATVBAHA121316797. Featured article of the issue.19. Ruter DL, Liu Z, Ngo KM, X S, Marvin A, Buglak DB, Kidder EJ, Bautch VL (2021). SMAD6 Integrates Endothelial Cell Homeostatic Flow Responses Downstream of Notch. Angiogenesis. 2021 May;24(2):387-398.
18. Ma H*, Liu Z*, Yang Y*, Feng D, Dong Y, Garbutt T, Hu Z, Wang L, Luan H, Cooper C, Li Y, Welch J, Qian L, Liu J (2021). Functional Coordination of Non-myocytes Plays a Key Role in Adult Zebrafish Heart Regeneration. EMBO Reports. 2021 e52901 https://doi.org/10.15252/embr.202152901. *co-first author.
2020
17. Wang L, Ma H, Huang P, Xie Y, Near D, Wang H, Xu J, Yang Y, Xu Y, Garbutt T, Zhou Y, Liu Z, Yin C, Bressan M, Taylor JM, Liu JD Qian L (2020). Downregulation of Beclin1 Promotes Direct Cardiac Reprogramming. Sci Transl Med 12(566): eaay7856.2019
16. Zhou Y*, Liu Z*, Welch JD, Gao X, Wang L, Garbutt T, Keepers B, Ma H, Prins JF, Shen WN, Liu JD, Qian L (2019). Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell 25 (1), 149-164. *co-first author.15. Ma H, Yu S, Liu X, Zhang Y, Fakadej T, Liu Z, Yin CY, Shen WN, Locasale JW, Taylor JM, Qian L, Liu JD (2019). Lin28a Regulates Pathological Cardiac Hypertrophic Growth through Pck2-Mediated Enhancement of Anabolic Synthesis. Circulation 139 (14), 1725-1740.
2018
14. Zhou Y, Alimohamadi S, Wang L, Liu Z, Wall JB, Yin C, Liu J, Qian L (2018). A Loss of Function Screen of Epigenetic Modifiers and Splicing Factors during Early Stage of Cardiac Reprogramming. Stem Cells International.2017
13. Liu Z*, Wang L*, Welch JD*, Ma H, Zhou Y, Vaseghi HR, Yu S, Wall JB, Alimohamadi S, Zheng M, Yin CY, Shen WN, Prins JF, Liu JD, Qian L (2017). Single Cell Transcriptomics Reconstructs Lineage Conversion from Fibroblast to Cardiomyocyte. Nature 551, 100-104 *co-first author.12. Zhou Y, Wang L, Liu Z, Alimohamadi S, Yin CY, Liu JD, Qian L (2017). Comparative Gene Expression Analyses Reveal Distinct Molecular Signature between Induced Cardiomyocytes and Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Cell Reports 20 (13), 3014-3024.
11. Liu Z, Chen O, Wall JB, Zheng M, Zhou Y, Wang L, Vaseghi HR, Qian L, Liu JD (2017). Systematic Comparison of 2A Peptides for Cloning Multi-Genes in a Polycistronic Vector. Scientific Reports 7: 2193.
2016
10. Liu Z, Chen O, Zheng M, Wang L, Zhou Y, Yin CY, Liu JD, Qian L (2016). Re-Patterning of H3k27me3, H3k4me3 and DNA Methylation during Fibroblast Conversion into Induced Cardiomyocytes. Stem Cell Research 16 (2), 507–518.9. Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin CY, Fu JD, Wang GG, Liu JD, Qian L (2016). Bmi1 Functions as a Critical Epigenetic Modulator during Early Phase of Direct Cardiac Reprogramming. Cell Stem Cell 18 (3), 382-95. Commentary: Herrer D and Benard A. Bmi1-Mediated Epigenetic Signature Acts as a Critical Barrier for Direct Reprogramming to Mature Cardiomyocytes. Stem Cell Investigation.
2015
8. Wang L, Liu Z, Yin C, Zhou Y, Liu J, Qian L (2015). Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5. JoVE (Journal of Visualized Experiments) e53426-e53426.2014
7. Wang L*, Liu Z*, Yin C, Asfour H, Chen O, Li Y, Bursac N, Liu J, Qian L (2014). Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming. Circulation Research 116 (2), 237-244. *co-first author. Editorial’s pick of the issue. Editorial: Muraoka N, Ieda M. Stoichiometry of Transcription Factors Is Critical for Cardiac Reprogramming. Circulation Research.6. Liu Z, F Zhao, He JJ (2014). Hepatitis C Virus (HCV) Interaction with Astrocytes: Non-Productive Infection and Induction of IL-18. Journal of Neurovirology 20 (3), 278-293
5. Liu Z, X Zhang, Q Yu, He JJ (2014). Exosome-Associated Hepatitis C Virus in Cell Cultures and Patient Plasma. Biochemical and biophysical research communications 455 (3), 218-22218.
2013
4. Liu Z, He JJ (2013). Cell-Cell Contact-Mediated Hepatitis C Virus (HCV) Transfer, Productive Infection and Replication and Its Requirement for HCV Receptors. Journal of Virology 87 (15), 8545-58.3. Park IW, Ndjomou J, Wen Y, Liu Z, Ridgway ND, Kao CC, He JJ (2013). Inhibition of HCV Replication by Oxysterol-Binding Protein-Related Protein 4 (ORP4) through Interaction with HCV NS5B and Alteration of Lipid Droplet Formation. Plos One 8 (9), e75648.20.